While new DNA sequencing technologies make it possible to better characterize tumors, they generate a large volume of data that must be interpreted.
GenerateReports is a bioinformatics platform for analyzing raw next-generation sequencing data in a clinical report that clearly summarizes the main sequencing results.
Its design allows:
- a retrospective analysis of biological samples as part of research projects
- interpretation of samples on a daily basis as part of a diagnostic activity
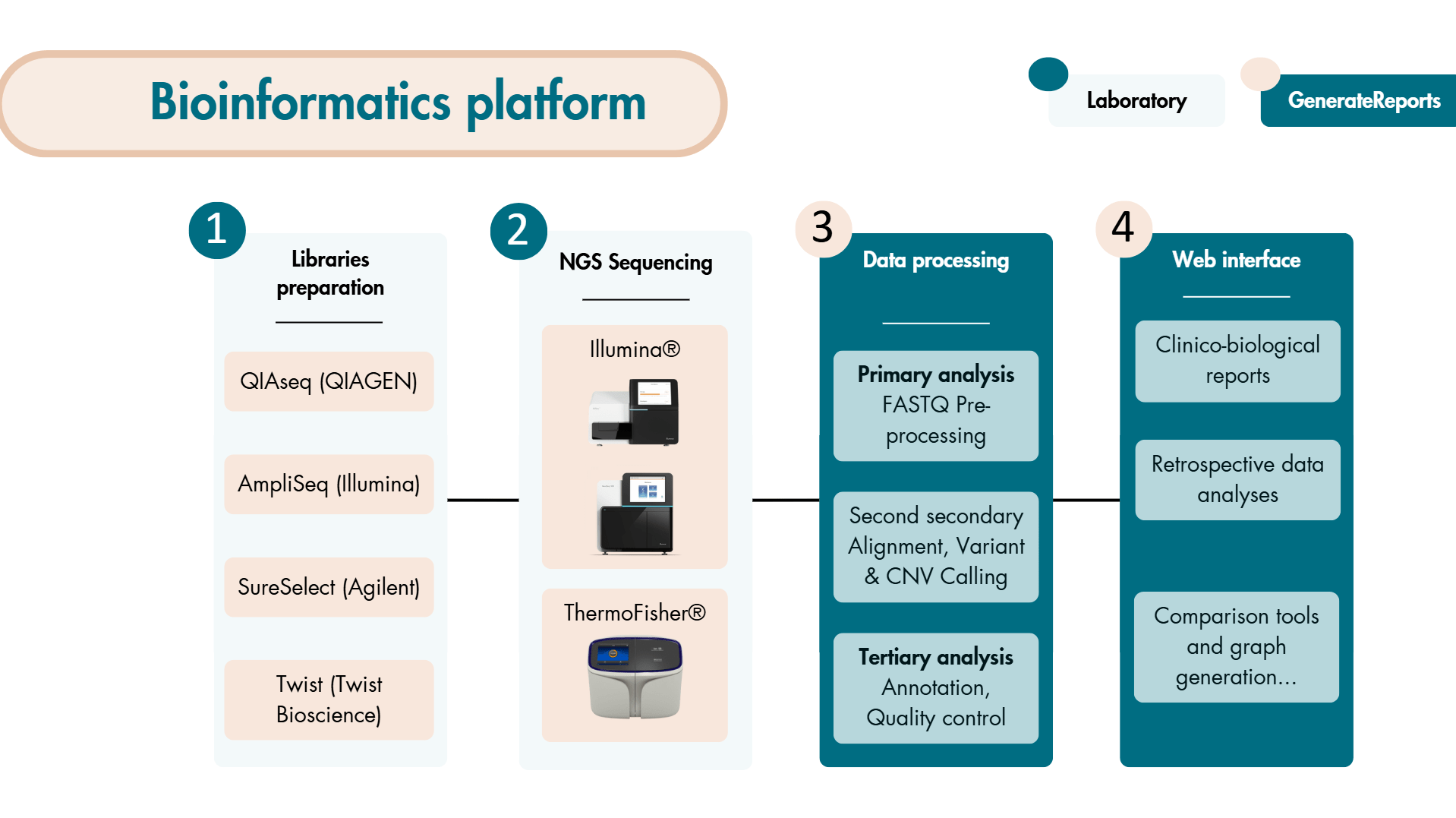
GenerateReports is above all a modular tool allowing you to create bioinformatics processing chains depending on the nature of the constructions of the sequencing libraries (targeted panels, exomes, capture, amplicon, etc.) and the biological questions asked (diagnosis/research, tumor/ctDNA sequencing, etc.).
it is accreditation assistance to the diagnosis of high-throughput sequencing (NGS) technology by enabling fine-grained control of bioinformatics pipeline versions and their dependencies.
GenerateReports integrates the notion of a list of variants to enable personalized bioinformatics analyses.
Sequencing report for each sample
For each sequenced sample, a detailed report is generated, providing a complete and transparent view of the results obtained. This report includes the following elements:
- Sample identification data
- Quality control on the scale of the Run
- Specific quality control of the sample
- The list of genetic variations detected and annotated
- Variations in gene copy number
- Experimental traceability and bioinformatics traceability information.

All data is also aggregated in a database allowing retrospective consultation of results at the following scale:
- of a sample or a variant
- of a complete project
- of a cohort of patients
- of an experiment
Variation lists can be created by biologists, making it possible to create documents, custom filters and certain automated analyses.
The database also allows the storage of external experimental validations to confirm the results obtained by high-throughput sequencing.
The software allows the insertion of data from various sequencing platforms.
| Library Compatibility | QIAseq (QIAGEN) AmpliSeq (Illumina) SureSelect (Agilent) Twist (Twist Biosciences) For other bookstores, contact us |
| Sequenced data type | Gene panels Whole Exome Sequencing (WES) Whole Genome Sequencing (WGS) |
| Variants detected | Single Nucleotide Polymorphisms (SNPs) Insertion & Deletion (INDEL) Mid-sized INDEL (~100b) Panel/exome/whole genome Copy Number Variant (CNVs) |
| Material compatibility | Sequencer Illumina® ThermoFisher ION S5 Sequencers |
| Supported data formats | FASTQ file |
| Databases used | 1000G ExAC ESP6500siv2 CG46 |
| Prediction scores used | ADA_score RF_score Score SIFT Score PolyPhen LRT MutationTaster MutationAssessor FATHMM PROVEAN VEST3 MetaSVM MetaLR MCAP CADD fathmm_MKL_coding GERP Interpro_domain |
Results obtained
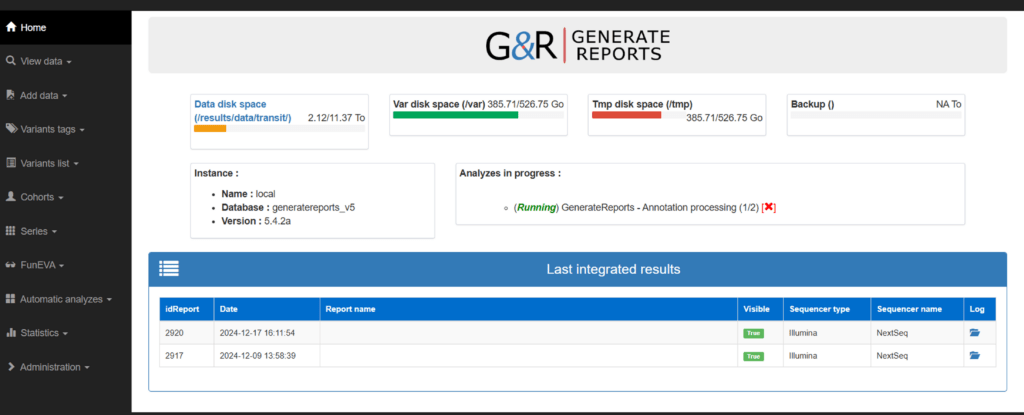
Access GenerateReports documents
Why are you asking me those information ?
By providing this information, you agree to be contacted so that we can best respond to your requests. This allows us to provide you with additional personalized information. Your data will be deleted upon simple request.
Publications related to GenerateReports
-
Somatic mutations of cell-free circulating DNA detected
PMID: 25749829

