Fusion transcripts are found in approximately 1/3 of soft tissue tumors and to date, the literature has described more than 140 fusions. They are often associated with a particular histological subtype and, consequently, with specific treatments. Thus, they constitute ideal molecular diagnostic markers.
The detection of these fusion transcripts is therefore essential for patient management.
The SarcomaFusion test test allows the detection of fusion transcripts identified in sarcomas in a single test.
This test is CE-IVD marked for diagnostic use in Vitro test, guaranteeing its compliance with European safety and performance requirements.
Here are some examples of detectable fusion transcripts associated with their pathology and possible treatments:
- Alveolar rhabdomyosarcoma
- Fusions PAX3::FOXO and PAX3::NCOA1
- Possible treatments
- chemotherapy
- pazopanib (PDGFa, VEGF-1)
- sorafenib (PDGFR, VEGFR, MAPK)
- crizotinib (MET, ALK, ROS1)
- temsirolimus (mTOR)
- cixutumumab (IGF-1R)
- radiotherapy
- Ewing sarcoma
- Fusions EWSR1::FLI1, EWSR1::ETV1, EWSR1::FLI1, EWSR1::FEV and other EWSR1 partners
- Possible treatments
- surgery
- chemotherapy
- radiotherapy
- CAR-Tcell therapy
- Epithelioid hemangioma
- Fusion ACTB::FOSB
- Possible treatments
- surgery
- chemotherapy
- radiotherapy
SarcomaFusion test is a unique, extremely robust and sensitive test intended for anatomopathologists and molecular biologists.
Based on ligation-dependent PCR technology (RT-MLPSeq), this test allows the detection of fusion transcripts, from a fresh, frozen or paraffin-embedded sample, among 140 possible in a single reaction.
The simplicity of the test allows the results of the analysis to be obtained in 24 hours from the tumor RNA. In addition, since the amount of tumor RNA required is very small, a needle biopsy is sufficient to obtain results.
SarcomaFusion contains a GAPDH control to ensure the success of the manipulation.
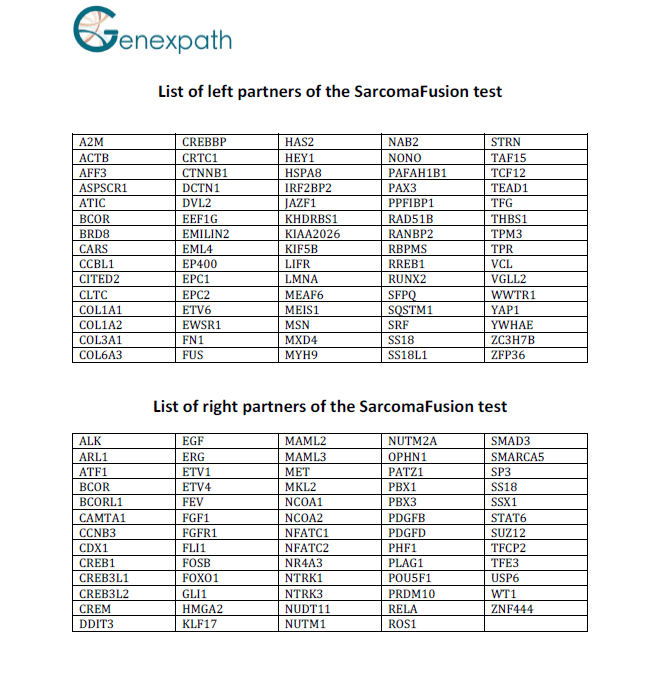
List of probes
The SarcomaFusion test contains probes specific to left and right partners of fusions described in the literature associated with sarcomas.
The key points of the protocol
In Vitro test
- 4 big steps
- 1/2 day including 1 hour to 1 hour 30 minutes of actual handling
- Sensitive thanks to short probes
- Optimized for FFPE samples
Sequencing
- Adapted to Illumina technology
- Possible to Sequencing with others
- Only 100,000 reads are needed per sample
Analyzed
- Increased specificity thanks to UMI
- Bioinformatics analyzes included
- Access to all raw data
| Cancer type | Sarcomas |
| Application domain | Fusion transcript detection |
| handling duration | ≃5h30 (without sequencing) |
| Actual working time | ≃1h-1h30 (without sequencing) |
| Sample types | Fresh, frozen or fixed and paraffin-embedded tissue biopsies |
| Input quantity | Between 50 and 500 ng of RNA in a volume of 2,5µL. |
| Reagents | Probes targeting over 140 fusion transcripts, barcodes, sequence primers |
| Material compatibility | Sequencer Illumina® |
| Bio informatique analyses | Access to the RT-MIS analysis platform included |
| Accessible data | Graphical representation of the detected fusion transcript, associated bibliography and complete raw data (csv) |
Kit composition
What the kit contains:
- RT-MLPA probe mix
- Genexpath barcodes
- Genexpath GAPDH control barcodes
- Sequencing primers
Available in 8, 16, 24 or 48 analyzes
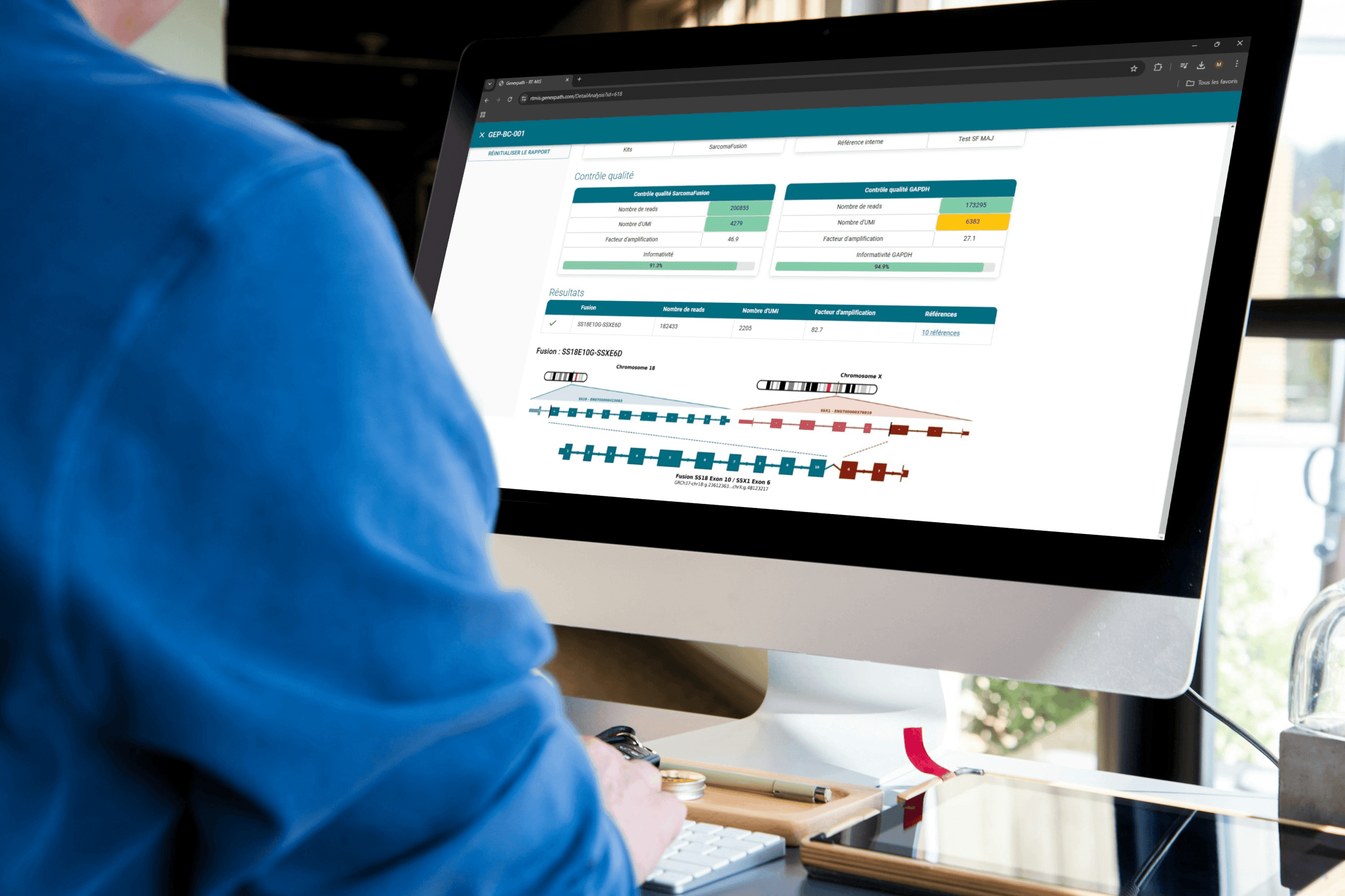
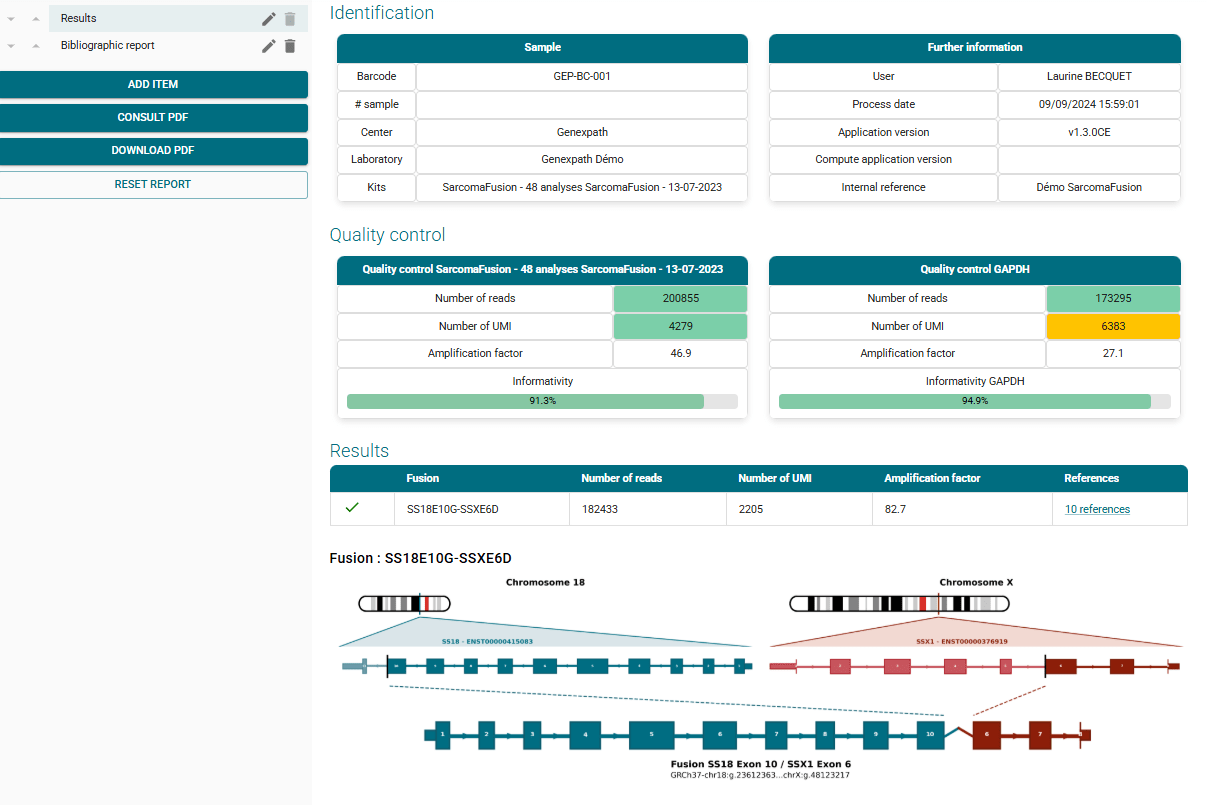
Results obtained
Once your FASTQ file is uploaded to the platform, RT-MIS performs demultiplexing and fusion transcript detection. Positive results are displayed as follows:
Access to SarcomaFusion's documentation
Why are you asking me those information ?
By providing this information, you agree to be contacted so that we can best respond to your requests. This allows us to provide you with additional personalized information. Your data will be deleted upon simple request.